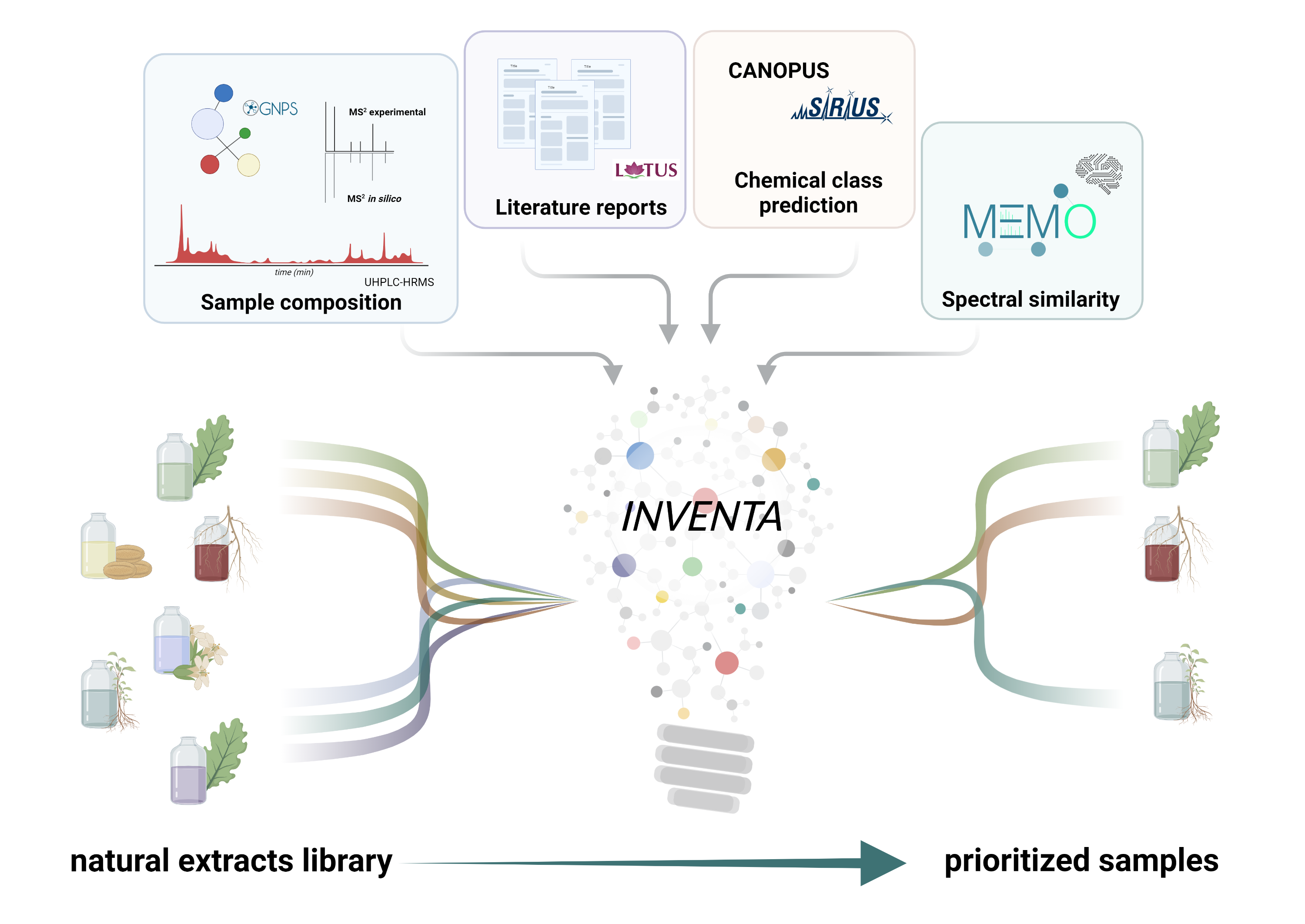
Inventa

a computational tool to discover chemical novelty in natural extracts libraries
Version 1.0
Graphical abstract
Description
Inventa calculates multiple scores that estimate the structural novelty potential of the natural extracts. It has the potential to accelerate the discovery of new natural produts.
The rank comes from the addition of four individual components:
The Feature Component (FC) is a ratio of the number of specific non-annotated features over the total number of features of each extract. For example, an FC of ‘0.6’ implies that 60% of the total features in a given extract are specific within the extract set and do not present structural annotations. Detailed FC calculation steps
The Literature Component (LC) is a score based on the number of compounds reported in the literature for the taxon of a given extract. It is independent of the spectral data. For example, an LC value of 1 indicates no reported compounds for the considered taxon. From this initial value (‘1’), fractions (ratio of reported compounds over the user-defined maximum value of reported compounds) are subtracted. The first fraction is related to compounds found in the species, the second one to those found in the genus, and the third one in the family. Detailed LC calculation steps
The Class Component (CC) indicates if an unreported CANOPUS chemical class is detected in a given extract compared to those reported in the species and the genus. A CC value of 1 implies that the chemical class is new to both the species (CCs 0.5) and the genus (CCg 0.5). Detailed CC calculation steps
The Similarity Component (SC) is a complementary score that compares extracts based on their general MS2 spectral information independently from the feature alignment used in FC, using the MEMO metric. This metric generates a matrix containing all the MS2 information in the form of peaks and neutral losses without annotations. The matrix is mined through multiple outlier detection machine learning algorithms to highlight spectrally dissimilar extracts (outliers). An SC value of ‘1’ implies the extract is classified as an outlier within the extract set studied.Detailed SC calculation steps
The combined score (adition of the four components) can be modulated acording to the user preference. The ouput consist of .tsv file with all the information generared along the final rank of the samples.
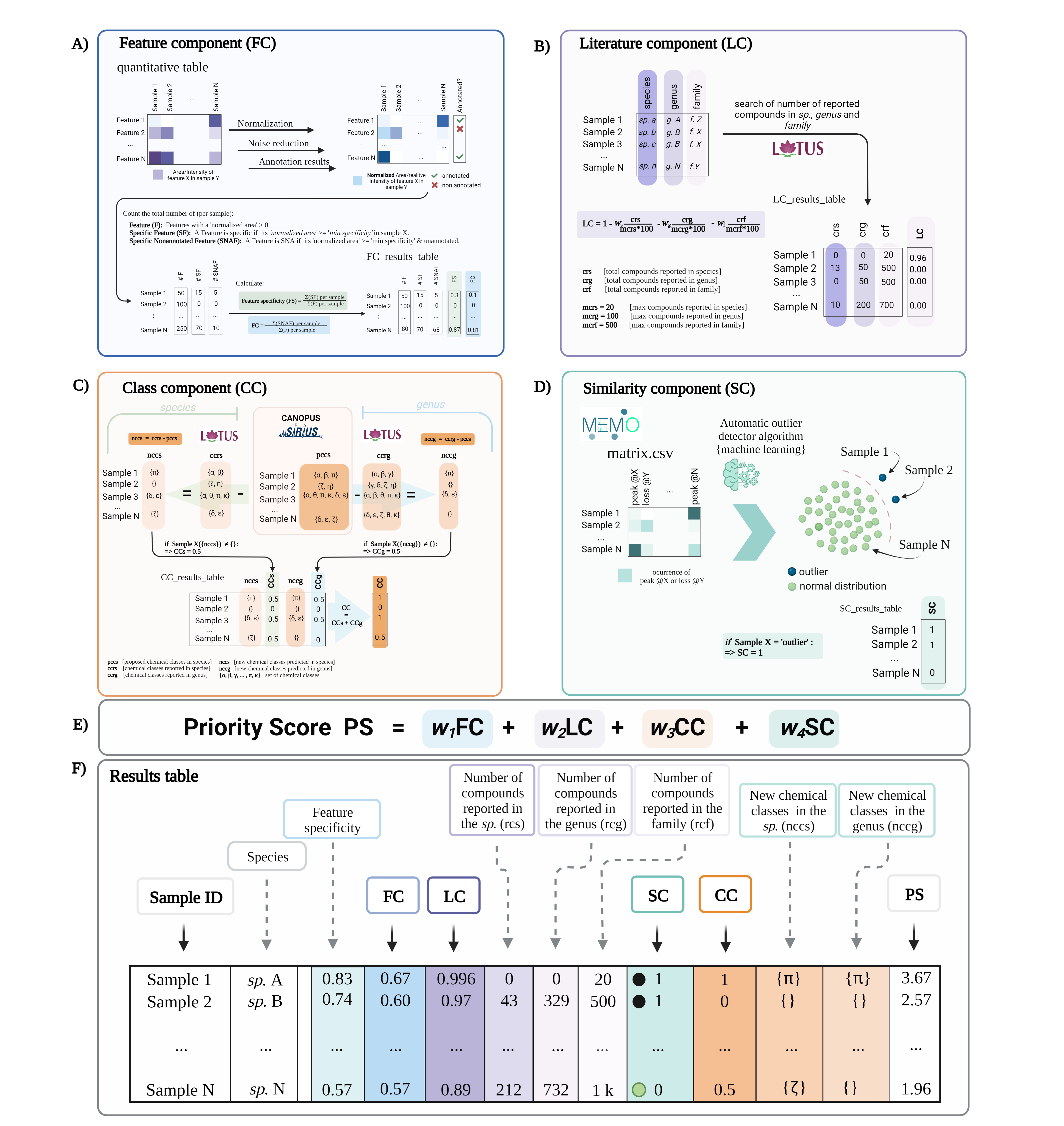
FIGURE 1. A conceptual overview of Inventa’s priority score and its components. (A) Feature Component (FC): is a ratio of the number of specific and unannotated features over the total number of features by extract. (B) Literature Component (LC): is a score based on the number of compounds reported in the literature for the taxon. It is independent of the spectral data. (C) Class Component (CC): indicates if an unreported chemical class is detected in each extract compared to those reported in the species and the genus. (D) Similarity Component (SC): Compares extracts based on their general MS2 spectral information though their MEMO vectors and automatic outlier detectors. This score is independent from any retention-time based alignment procedure and complementary to FC. (E) The Priority Score (PS) is the addition of the four components. A modulating factor (wn) gives each component a relative weight according to the user’s preferences. The higher the value, the higher the rank of the extract. (F) Results Table is a resume of individual calculation components and results.
Table of contents
1. Installation
2. Getting started
3. Configurations and running
4. Publication images
Credits
Images were created by Luis Quiros-Guerrero using bioRender (© BioRender 2022)
Copyright and license
Code and documentation copyright 2011–2022 the authors. Code released under the MIT License.